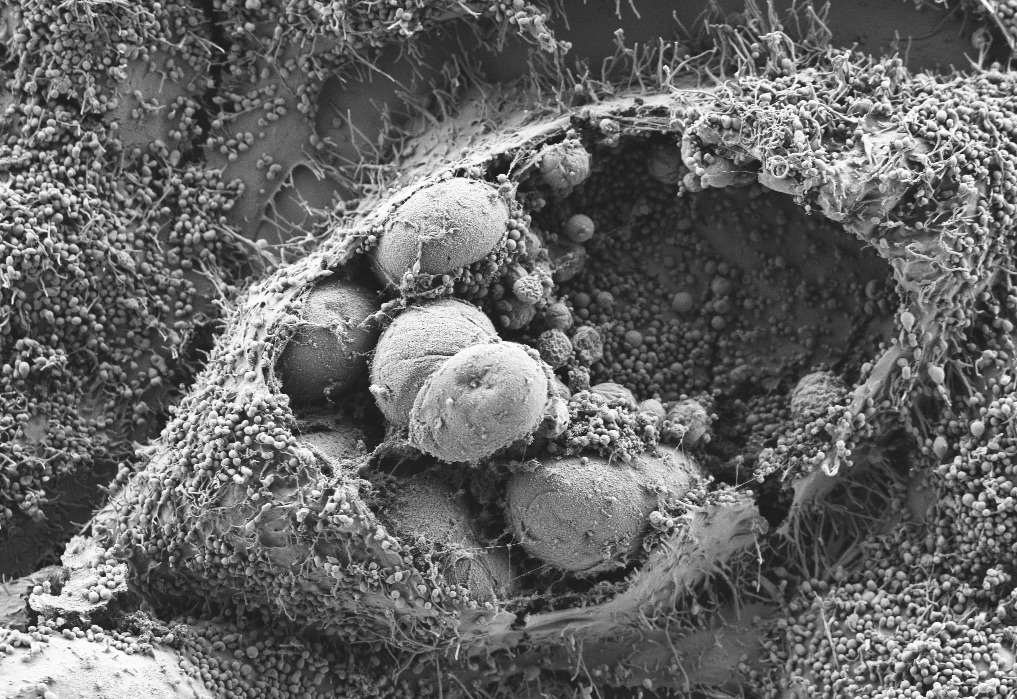
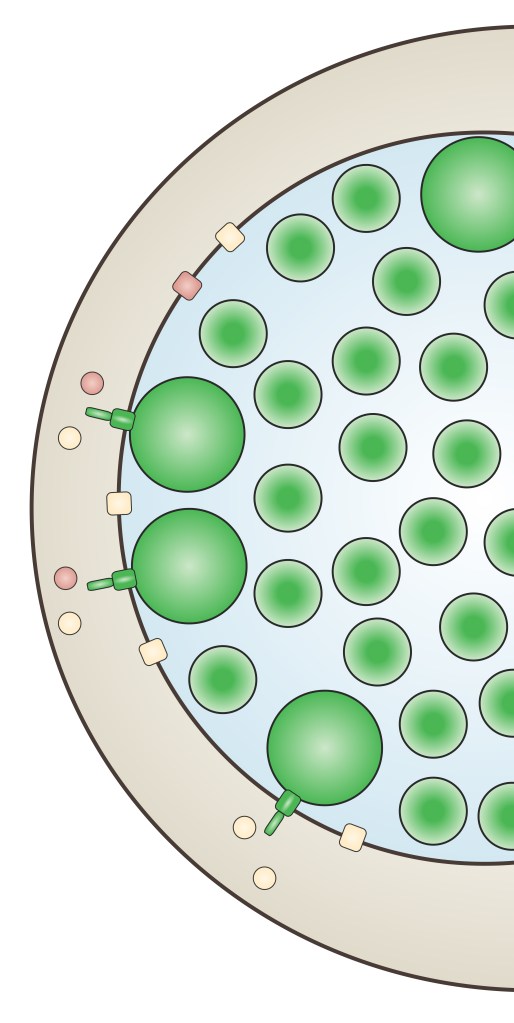
We study the pathogenesis of human chlamydial infections
The bacterium Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infection (STI) throughout the world. Chlamydia infection rates are high and continue to rise. Prior to COVID, Chlamydia infections were the most commonly reported infection in the US. Chlamydia disproportionately affects young women and people from disadvantaged backgrounds for the development of severe reproductive outcomes. While effective treatments exist, there is no preventative vaccine, and much remains to be learned about how this pathogen causes disease in humans.
The goals of our research are to define the critical bacterial and host factors responsible for Chlamydia infections and chronic clinical outcomes.